 Why-O-Why has this taken so long?
Why-O-Why has this taken so long?
Finally we have a preprint paper assessing SARS2 as if it might have been an engineered product from a laboratory. We now know it was very likely a lab product and we can probably even name the tools that were used to tweak it.
The virus appears to be too clean, lacking in the noise that all its wild type cousins have. Random evolution in bats and pangolins just doesn’t work like this. SARS2 has the unmistakable fingerprint pattern of a virus that not-so-coincidentally is perfectly suited to being manipulated with two of the most common laboratory enzymes available available at a biolab near you for $150. (Shop here, here, or here).
The authors stress that even though their results strongly suggest this virus was a “synthetic” virus, that doesn’t tell us whether it was intended to harm, or was released deliberately. But their results do cast a very different light on the rush to declare the wet markets were to blame, and the too-fast calls, based on no evidence, that anyone who said otherwise was a conspiracy theorist. This kind of analysis could have been done in Feb-March 2020 and it wasn’t. How fast would nations have slammed the borders shut if they thought this was a lab accident?
“This has been an incredible project. Yet, for obvious reasons, this is the saddest paper I’ve ever written.” — Alex Washburne
hat tip to Matt Ridley
*UPDATE added at the bottom as the debate over this paper progresses.*
So Valentin Bruttel, Alex Washburne, and Antonius VanDongen looked for evidence that the virus was manipulated to suit two of the most common gene-slicing tools for sale — called BsaI and BsmBI . As Alex says, when you walk into the store to buy enzymes, these are the top recommendations. What they found were that the target cutting sites for these were placed quite neatly separated, to produce workable size fragments of a similar size. They also found that important genes were not split — things like the key RBD code were neatly contained in one piece, rather than cut in the middle. (RBD means the Receptor Binding Domain — the key bit that sticks to ACE2). When they compare SARS2 to 37 other types of coronaviruses the mutations that produced these new target codes were conveniently “silent” mutations. They were neutral spelling changes in the code that don’t alter the final meaning. For example both CTC and CTA code for the same amino acid. So we can flip between the two and the end product made from this will be the same*. But change an ATG to a GTG and the shape of the protein made will be different. Obviously if a human brain was managing the code for some desired effect they would select the “silent” kind of mutations to either create new target sites or remove unwanted target code in the wrong spot.
This is a long post, but the era of bioweapons and genetic-engineering accidents is upon us whether we like it or not. These researchers conclude the SARS2 virus was likely synthetic but even if it wasn’t and someone comes up with a realistic criticism of the paper — the SARS2 virus could clearly have been made easily in many labs. Clearly we need to talk about this!
From the abstract:
Both the restriction site fingerprint and the pattern of mutations generating them are extremely unlikely in wild coronaviruses and nearly universal in synthetic viruses. Our findings strongly suggest a synthetic origin of SARS-CoV2.
To understand how genetic engineering works we need to know the basic tools
Once we know the basic gene editing techniques we can see how well adapted the virus is for lab work. So here is a ten-second instruction kit for genetic engineering. DNA is made a four letter code A, C, G, and T. It has two strands: the active code and the mirrored “back up” copy (something the computer coders will appreciate). Thanks to chemistry the A’s always pair with T’s, and the C’s pair with G’s. So, for example, the sequence of GGAA will “glue” itself to CCTT on the opposite paired strand. Our DNA is the ultimate chemical velcro. The chain of paired bases makes up the famous double helix.
We have a lot of off-the-shelf tools to slice up that double helix code, and each of these tools cuts only a certain sequence, say CGTCTC, which leaves handy “sticky ends” that can be glued back together. This short sequence will stay embedded in the code after the edit, which is a clue, but not much of one on its own, since these sort of short sequences can occur randomly too.
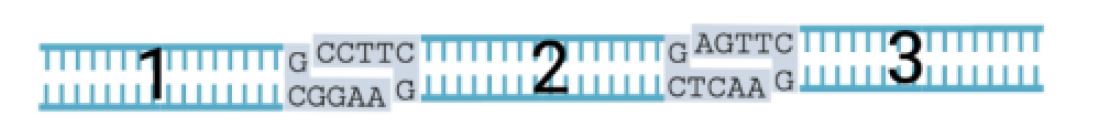
This is code ready to be cut into 3 parts with “sticky ends” that will reassemble back onto themselves (to loop) and then later reassembled back in a line. There are unique “sticky ends” so the loops only pair back to the right partner in the right order.
But the lack of these target sites is also a clue. In wild viruses these “target sites” will occur randomly in the wrong spots. So the first task of serious genetic engineers is to remove the badly placed target sites — if they don’t do that, the enzymes will chop up the DNA into odd sized fragments, too big and too small to work with. The code needs to be “clean”.
Hmm? Those fragment sizes in SARS2 are oddly “perfect”…
[EDITED] Here’s a graph of the number of fragments made from 70 different wild viruses which were chopped at all the wild target sites.The number of fragments is graphed against the size of the longest piece. The wild viruses produce the grey curve with a kind of random distribution of cutting sites. But synthetic viruses fall in the red box where 5 – 8 fragments of similar size is easier to work with.
Washburne explains this graph on his blog. To get the neat log curve they took 70 wild viruses and cut them up with 200 enzymes in 1,000 enzyme pairs. Engineered viruses end up in the red box. Wild viruses fall on the grey curve.
Genetic engineers need to play with nice medium length fragments
The basic technique for genetic engineering is below in the diagram. The virus is chopped up into separate bits. Each bit has a matching end that pairs up on itself to form a loop, which is easy for people to work with. Then after we have played games with the separate loops we can cut them open at the same target marks, put all the loops back together, and they will reassemble into the right order in the long chain. To make this work the fragments need be a workable length, and each “split” is unique so the chain will only form in the right order.
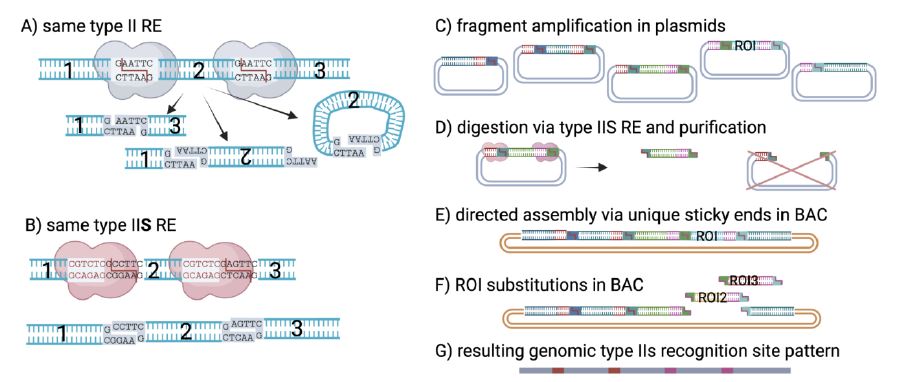
So we chop up DNA, roll it into little circles we can work with. Make the changes we want and the open the circles and reassemble them into one long chain of code.
It’s worth knowing that while SARS is a single strand RNA virus, to play games with it we convert it into DNA and use our DNA engineering tools. This is because in the world of chemistry — RNA is like a cheap photocopy of DNA. The molecules are nearly the same, but it has only one strand, and it’s so unstable and sticky that it’s very hard to work with. To engineer RNA, the lab techs convert it to DNA, then play games, then convert the end product back to RNA (click to expand the diagram of the process above). This is especially important with a large virus like the SARS-Cov-2 virus which has nearly 30,000 bases. (That’s one long strong of letters like… ACGTCCTGC… up to 30,000 “letters” in a row.)
So let’s imagine the target sites were placed randomly by mutation inside bats in a cave…
This below is an evolutionary branched tree of corona viruses with two kinds of SARS2 in the top two rows and compared to 37 other corona viruses below. As you can see, for some reason the bats that SARS2 came from have cleaned up the target sequences. Clever bats. The red and green dots are the target sites for our favorite genetic engineering tools. The spacings in the SARS 2 rows are oddly ideal. This is part of the fingerprint of a laboratory made virus.
Like wow: This table blew me away…
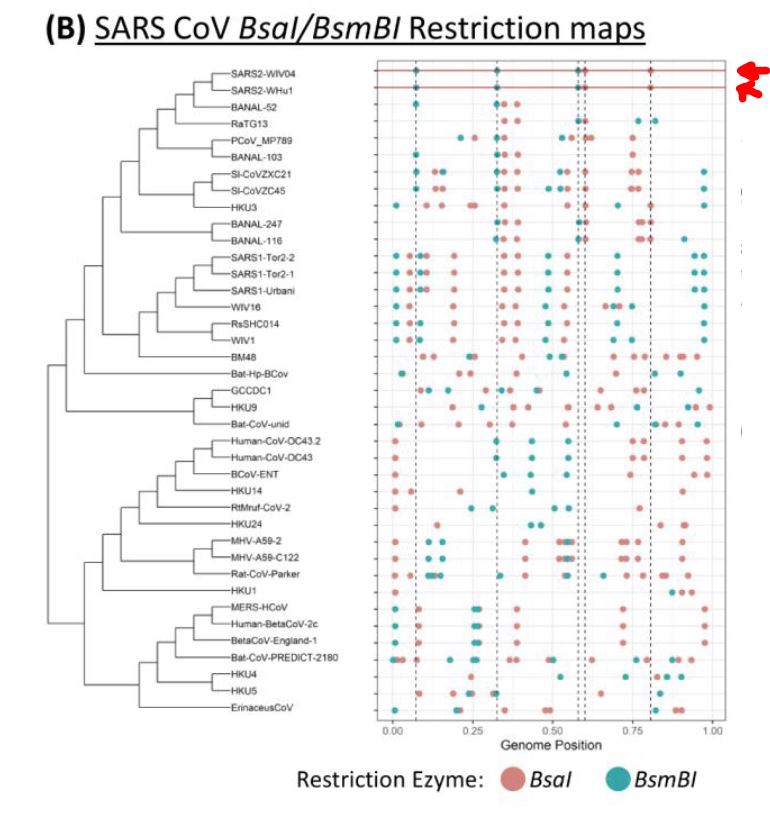
Note the striking even spacing of the cutting target sites in the top two SARS rows compared to the random pattern in all the other Coronaviruses.
There’s more to this to be a full fingerprint pattern of a lab virus:
On his blog Alex Washburne‘s describes a pattern that marks a virus as “lab made”. An “infectious clone” means a synthetic clone of a wild virus. It not only needs nicely spaced markers for enzymes available by mail order, but also the “sticky ends” must be unique so it can be reassembled in the right order, and the mutations must be silent, and the genes need to be contained neatly on single piece.
The Z-scores for 5 – 7 fragments suggest this was “more likely engineered”.
Your eyeballs knew it anyway…
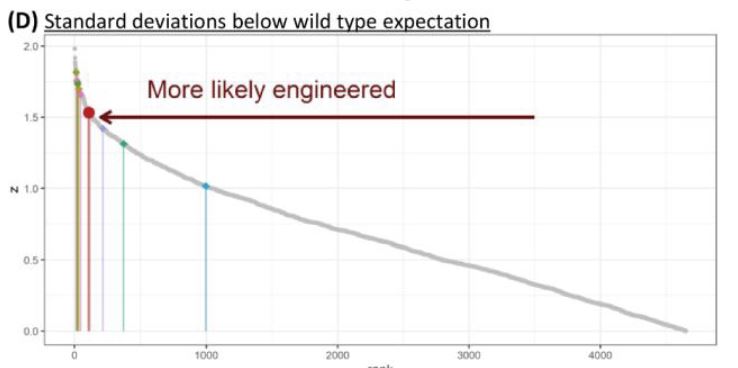
Caption: A ranked plot of z-scores for all digestions creating 5-7 fragments, the idealized range for a CoV reverse genetic system. z-scores measure the standard deviations below the wild type expectation, correcting for the number of fragments. SARS-CoV-2 appears more likely to have been engineered for IVGA than several known CoV reverse genetic systems.
So this then is the final SARS2 compilation of fragments assembled
Here’s the assembly plan for the virus that held up the world. The lowest layer in the diagram below is the old SARS1. We can see the placement of target sites for the two common laboratory enzymes (marked in red). As we go up each layer (to a new different virus) things are cleaner than the layer before. The topmost layer is SARS2 itself. The RBD and FCS (Furin Cleavage site) are marked. Click to make this bigger.
This is the covid virus, at the top, in segments. Start with wild coronaviruses at the bottom, they have random spaced target sites. But the sites in Covid are neatly spaced just where an genetic engineer would want them to be.
All the clues point to a lab created virus:
What are the odds a million bats typing on typewriters could create this?
Washburne tries to calculate the odds of this pattern occurring naturally.
It turns out, the sticky ends produced by BsaI/BsmBI digestion of SARS-CoV-2 are all unique, non-palindromic, and all contain at least one A or T – all criteria either required or recommended for in vitro genome assembly.
It also turns out, the mutations separating SARS-CoV-2 BsaI/BsmBI sites from those of its close relatives are all silent. About 84% of mutations between SARS-CoV-2 and its two closest relatives (BANAL-20-52 and RaTG13) are silent. There are 14 distinct mutations separating SARS-CoV-2 BsaI/BsmBI sites, and all of them are silent. There’s a ~9% chance that 14 randomly drawn mutations are all silent.
Compared to the rest of the genomes, we found a significantly higher rate of silent mutations within BsaI/BsmBI sites. Between BANAL52 and SARS-CoV-2, there is a 5x higher rate of silent mutations within BsaI/BsmBi recognition sites than the rest of the genome (P=0.004). Between RaTG13 and SARS-CoV-2, there’s a whopping 8x higher rate of silent mutations with 1/100 million odds of seeing as high or higher concentration of silent mutations within the BsaI/BsmBI restriction sites.
The odds of meeting any one of these criteria vary, from 1%-0.07% of having such a small maximum fragment length to 1/250 to 1/100 million odds of having such high concentration of silent mutations within BsaI/BsmBI recognition sites. The odds of meeting every single one of these criteria are even smaller. Much smaller.
As a result of this analysis, we theorize that SARS-CoV-2 was assembled in a lab via common methods used to assemble infectious clones pre-COVID.
It looks just like a common research product from a lab:
SARS could be an experiment that escaped, or it could be a bioweapon. This research does not differentiate, but obviously, if it’s a lab leak, that changes everything, and it also would have changed everything if this had been known in February 2020:
A synthetic origin of SARS-CoV-2 doesn’t mean there was malicious intent. In fact, the location of BsaI/BsmBI site appear almost chimeric: some are shared with close relatives and others are shared with distant relatives, making us hypothesize that perhaps researchers just wanted to make chimeric viruses within this clade of relatively unstudied CoVs. Again, this is a common research project, much like that conducted at Boston University recently, and it is often done with noble intentions of learning about genotype-to-phenotype relationships and even preemptively designing vaccines against viruses that are most-likely to cause a pandemic. That would be tragically ironic if proven true.
Many labs could have done this kind of work says Washburne on his blog. The world needs to wake up to the ease with which this kind of dangerous work can be done:
We don’t identify who constructed the virus. Many people in the world could do this, although the origin of this outbreak in Wuhan does narrow the range of suspects considerably. The technology used to make infectious clones is relatively cheap, especially compared to making an atom bomb. Even if our theory is rejected by later tests, the ease of these experiments should scare the shit out of all of us enough to start talking about global biosafety.
We need to urgently discuss of how to prevent accidents
Perhaps all labs should have signature markers inserted in their work?
In addition to, I don’t know, regulating which sequences you can purchase online, we may also want to require some identifiability of chimeric experiments. As silencers for guns are illegal, we may be wise to require all chimeric viral research have clearly identifiable sequences that help us identify right away who did it and what was done, as such information may be relevant for preventing a lab accident from turning into a full-blown pandemic.
Washburne writes in beautifully dispassionate tones — always trying to figure out why he might be wrong:
As a scientist conducting this research, I did my best to ensure our methods were reproducible, our statistics conservative, and our presentation honest. We discovered SARS-CoV-2 was unusual, knew the massive stakes of our finding, and set out to disprove our hypothesis by looking closer at sticky ends, silent mutations, and analyses of the evolution of CoVs. Any one of these tests could’ve rejected our hypothesis and the world would never have seen this paper. We sought peer-reviews from world experts at every leading institution we could connect with, and we asked them to shake down our results. The pre-print is not exactly “not peer-reviewed”, as it is the product of rolling feedback from world experts and we did our best to incorporate all of their feedback, test all of their proposed tests, and include all of the limitations they identified in our manuscript.
The authors stress that “synthetic” means synthesized, and not necessarily a bioweapon, or “Gain of Function”. But even if SARS2 wasn’t a bioweapon we need to discuss these tools and techniques. There are other aspects outside this work that are relevant to the bioweapon question, and we’ll discuss those another day. For the moment, the citizens of the free world need to learn enough about genetic engineering to discuss what kind of world we want to live in.
___________
UPDATE: It’s great to see commentary on this paper starting. Alex Crits-Christoph has attacked the paper on a twitter thread, which Washburne has already responded to. The former points out that the the pattern in other enzyme targets in SARS2 are not unusual and claims this is cherry picking, but if a team did engineer the virus to suit one or two enzymes I don’t see why we wouldn’t see what Crit-Christoph finds? Washburne replies they didn’t cherry pick they just started looking at two of the most popular enzyme tools. They also used a long list of criteria.
Other critics are pointing out the the Golden Gate Assembly model would remove the “scars” (or traces) of the Golden Gate sequences. But are these scar or coding elements removed later, only at the end? Tony vanDongen (author) says it’s correct the process can be scarless with target sites removed, but then they can’t be used again. He implies SARS2 is a work in progress — “The BsaI/BsmBI fingerprint in SARS2 is indicative of a design where you could test many different iterations of the virus. Disassemble and reassemble the genome at your heart’s content.” That suggests SARS2 may have been a workhorse used to test many things in a lab?
EDITED: Washburne notes that “The most important tweak is that the exact method used to assemble CoVs isn’t best termed “Golden Gate“ but “Type IIS directional assembly”. So “Golden Gate was removed from the introduction paragraphs.
____________
REFERENCES
Valentin Bruttel, Alex Washburne, Antonius VanDongen (2022) Endonuclease fingerprint indicates a synthetic origin of SARS-CoV-2,
Alex Washburne’s blog and his twitter thread.
*Silent mutations turn out to still make a difference sometimes. The amino acid is the same but the protein may fold at a different rate to a slightly different shape. Dang life is complicated.